One-step TUNEL In Situ Apoptosis Kit (Green, FITC)
Detection principle
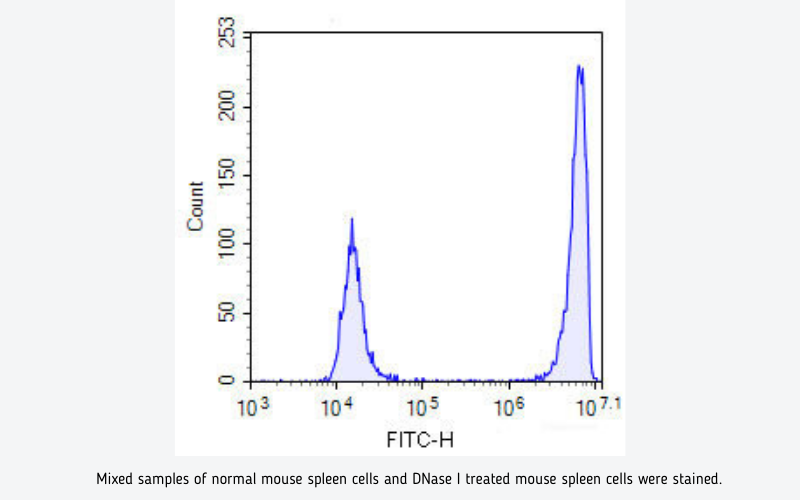
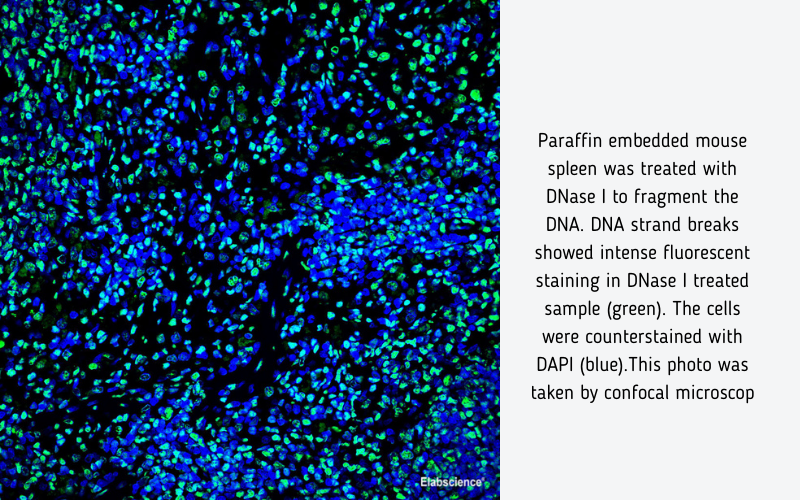
When cells undergo apoptosis, specific DNA endonucleases will be activated, cutting the genomic DNA between the nucleosomes. The DNA of apoptotic cells is cleaved into multimers of 180~200bp fragments, corresponding to the oligonucleosomal size. Therefore, the DNA of apoptotic cells typically migrates as a ladder of 180~200bp on an agarose gel. The exposed 3'-OH of the broken DNA can be catalyzed by Terminal Deoxynucleotidyl Transferase (TdT) with fluorescein labeled dUTP, which can be detected with fluorescence microscope.
General Information
| Application | DNA Fragmentation |
| Detection method | Fluorometric Method |
| Sample type | Paraffin section, frozen section, cell slide |
| Assay time | 3 hours |
| Detection instrument | Fluorescence Microscope |
| Dye Type | FITC |
| Ex/Em (nm) | 490/520 |
| Channel set | FITC |
| Filter set | FITC |
| Other reagents required | 4% Paraformaldehyde(E-IR-R113),0.2% Triton X-100(E-IR-R122),PBS(E-IR-R187),ddH2O,Mounting solution with anti-fluorescence quencher(E-IR-R119),Xylene,Ethanol |
| Storage | -20°C, shading light |
| Shipping | Ice bag |
| Expiration date | 12 months |
Application
Apoptosis/DNA Fragmentation
Operation Guide
This video is about how to operate Elabscience® One-step TUNEL (Cell Slide) experiment. For each reaction, there is a demonstration, so that the experimenters can better understand the TUNEL process.
Procedure:
Cell Slide → Fixation → Washing →Permeabilization → Washing → Equilibrium → Labeling → Washing → Nuclear staining → Washing → Mounting → Analyze sample
For research use only.
Ordering Information
| Cat.No. | Pack Size |
| E-CK-A320 | 20 assays / 50 assays / 100 assays |